Other proteins instead dynamically exchange with those in solution soft corona56 Nevertheless our knowledge. This protein is at the centre of vaccine development because they trigger an immune response in humans.

Hidden States Of The Covid 19 Spike Protein Eurekalert Science News
21062021 Molecular dynamics methods are playing an increasing role in the research of protein-DNA recognition.

Protein corona molecular dynamics. The simulations analyzed with VMD revealed that transport factors are dotted rather regularly on their surface with spots that bind to the brushes of nuclear pore proteins. Systematic molecular dynamics simulations further provided molecular and structural details for the observed differential effects of proteins on IAPP amyloidosis. After interaction the two proteins bind together.
In simulations of a single protein with a PS particle proteins eventually. Molecular dynamics simulations based on relevant crystallographic structures using NAMD provided a comprehensive picture on the passage mechanism as reported recently. 01092002 Molecular dynamics simulations of biological macromolecules have provided many insights concerning the internal motions of these systems since the first protein was studied over 25.
Understanding the corona formation process is crucial in predicting nanoparticle behavior in biological systems including applications of nanotoxicology and development of nano drug delivery platforms. Animation showing how the coronavirus spike protein changes its shape just before binding to human cell receptor. In the first phase proteins rapidly bind to the free surface of nanoparticles leading to a metastable composition.
The electron microscopy images also confirmed that protein corona reduced the cellular morphological damage by limiting GO penetration into the cell membrane. Our cell viability and cellular uptake experiments showed that protein corona decreased cellular uptake of GO thus significantly mitigating the potential cytotoxicity of GO. Nanoparticle-protein corona complex formation involves absorption of protein molecules onto nanoparticle surfaces in a physiological environment.
05012021 The trajectory obtained from the molecular dynamics simulations was analyzed to identify critical residues at the interface between spike protein and ACE2. The first molecular dynamics simulation of a protein was reported in 1977 and consisted of a 92-ps trajectory for a small protein in vacuum 4. These proteins equip ENMs.
19072019 The results discussed in the light of the controversial experimental data reported in the current experimental literature have provided a detailed description of the i recognition process ii number of proteins involved in the early stages of corona formation iii protein competition for AuNP adsorption iv interaction modalities between AuNP and protein binding sites and v protein. Like a balloon on a string the spikes appear to move on the surface of the virus and thus are able to search for the receptor for docking to the target cell describes Jacomine Krijnse Locker group leader at the. 19082020 At the start of a Covid-19 infection the coronavirus Sars-CoV-2 docks onto human cells using the spike-like proteins at its surface.
11062021 The protein corona is a layer of proteins that spontaneously adsorbs to the surface of engineered nanomaterials ENM when exposed to biological systems 1 2 3 4 5 6. We show that the coronal layer structure is responsive to the dielectric constant of the medium and that the mobility of the polymer surfactant molecules is significantly hindered in the solvent-free state providing a basis for the origins of retained protein dynamics in these novel biofluids. 10052005 Molecular dynamics simulations provide links between structure and dynamics by enabling the exploration of the conformational energy landscape accessible to protein molecules 1 3.
During the second phase continuous association and dissociation of protein molecules with nanoparticles slowly changes the composition of the corona complex. 14042020 Enhanced detailed simulations of such molecular dynamics will provide a complete picture of proteins structural changes as well as how they bind to angiotensin-converting enzyme 2 the specific human cell receptor. 07082020 The human proteins which coats inside of the nose and the cells of lungs are mainly can interact with the spike protein of the SARS-CoV-2 virus.
Stabilities of the trimeric form of the spike protein complexed with ACE2 and fluctuation of residues in the simulations were examined using root mean squared deviation RMSD of Cα atoms and root mean square. 31052013 Results The model presented in this paper exhibits two phases of corona complex dynamics. These methods have been used to study the specific and non-specific binding of proteins to nucleic acid the kinetics and thermodynamic behavior of the binding process and the regulatory mechanism of the chemical modification of DNA on the.
Nowadays it is generally accepted that part of the proteins in the corona can remain for a relevant time on the NP surface hard corona4 possibly preventing the adsorption of other molecules. 18082020 By combining molecular dynamics simulations and cryo tomography the team identified the three joints hip knee and ankle giving the stalk its flexibility. This study facilitates our understanding of the fate and transformation of IAPP in vivo which are expected to have consequential bearings on IAPP glycemic control and T2D pathology.
That the protein corona is composed of an inner layer of tightly bound molecules called the hard corona and an outer layer of soft corona consisting of molecules only loosely attached to the NP surface1011 Whereas molecules in the soft corona undergo dynamic adsorptiondesorption cycles and thus only. In the product form the spike protein of CoV-2 changes its shape and causes the human receptor cell to engulf the virus. Molecular Dynamics of Proteins ATPase a molecular motor that synthesizes the bodys weight of ATP a day A ternary complex of DNA lac repressor and CAP controlling gene expression H-bond energy kcalmol 0 - 40 Fibronectin III_1 a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis.
All-atom molecular dynamics simulations of plasma proteins human serum albumin fibrinogen immunoglobulin gamma-1 chain-C complement C3 and apolipoprotein A-I adsorbed onto 10 nm sized cationic anionic and neutral polystyrene PS particles in water are performed.

An In Silico Study On Selected Organosulfur Compounds As Potential Drugs For Sars Cov 2 Infection Via Binding Multiple Drug Targets Sciencedirect
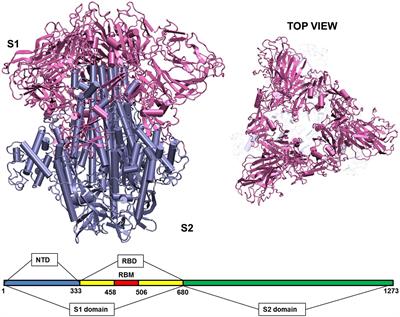
Computational Approaches To Understand Molecular Mechanisms Of Sars Cov 2 Frontiers Research Topic
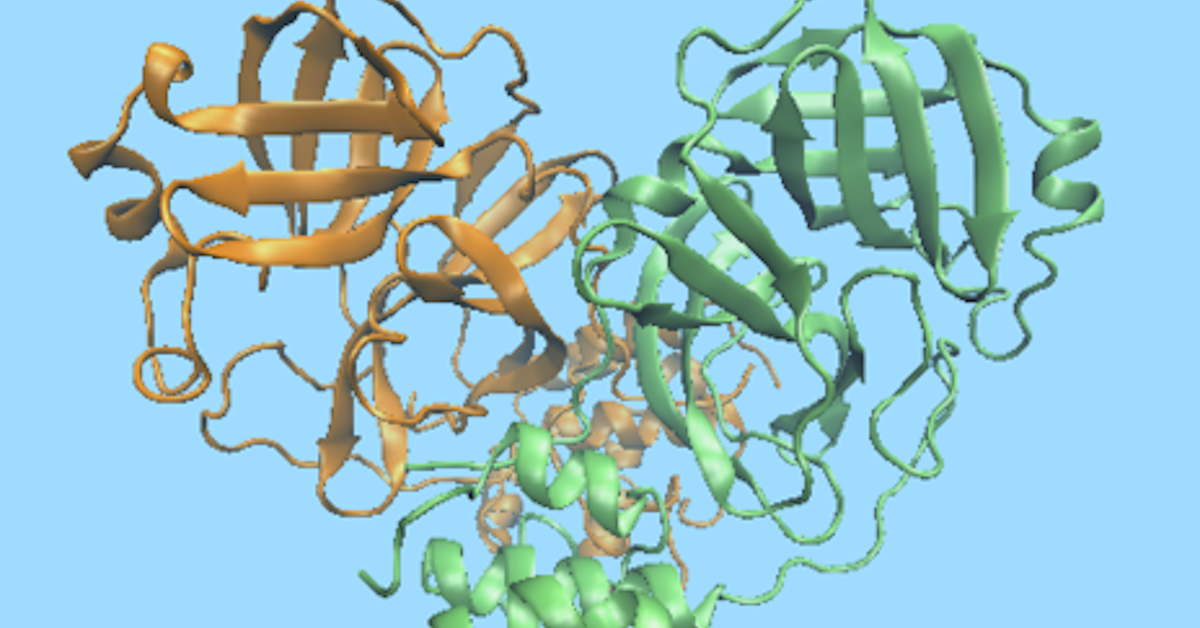
Exploring The Molecular Dynamics Of The New Coronavirus Riken
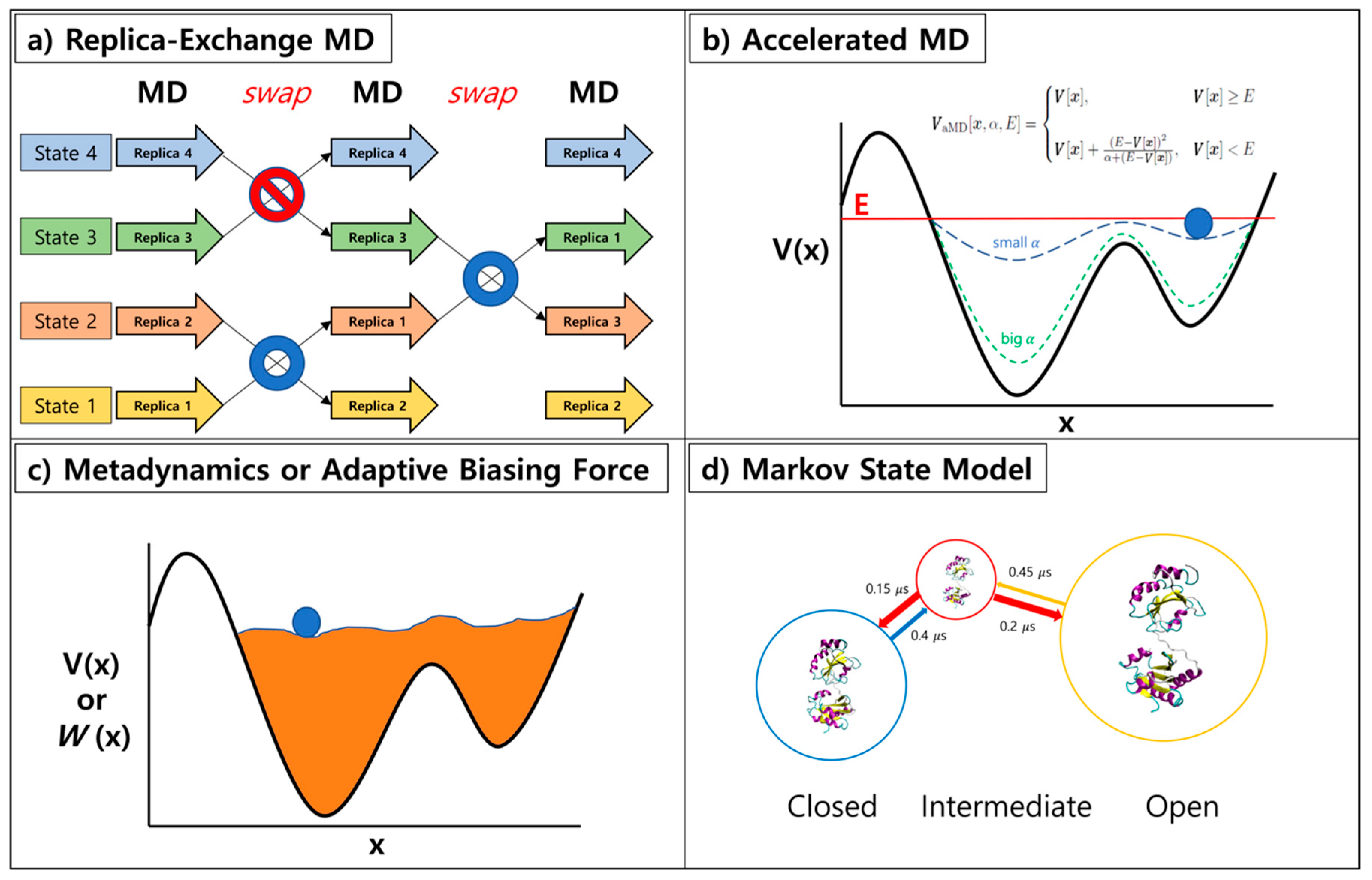
Ijms Free Full Text Advances In Molecular Dynamics Simulations And Enhanced Sampling Methods For The Study Of Protein Systems Html
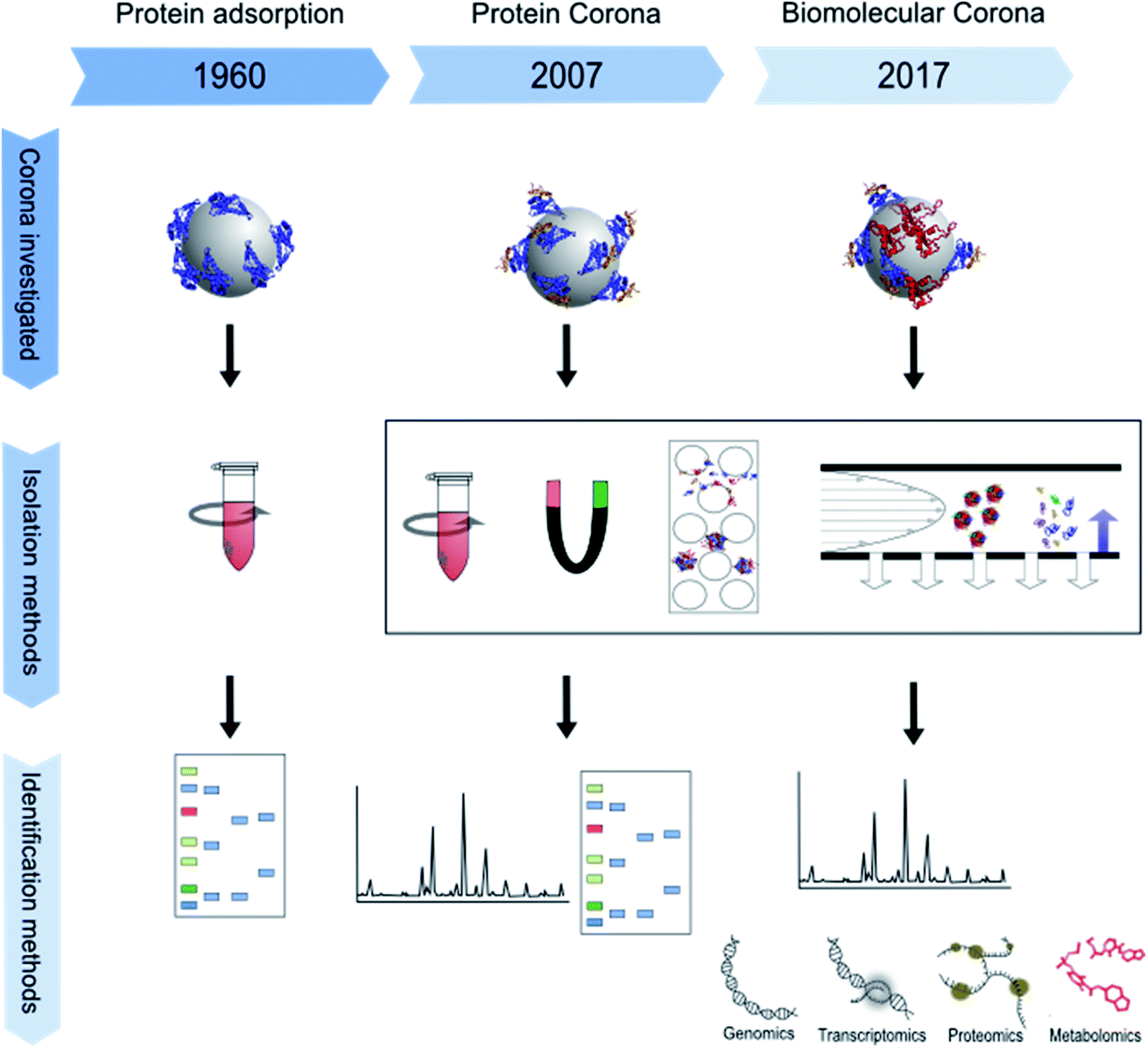
Isolation Methods For Particle Protein Corona Complexes From Protein Rich Matrices Nanoscale Advances Rsc Publishing Doi 10 1039 C9na00537d

Towards A Molecular Level Understanding Of The Protein Corona Around Nanoparticles Recent Advances And Persisting Challenges Sciencedirect

Sars Cov 2 Covid 19 Structural And Evolutionary Dynamicome Insights Into Functional Evolution And Human Genomics Journal Of Biological Chemistry
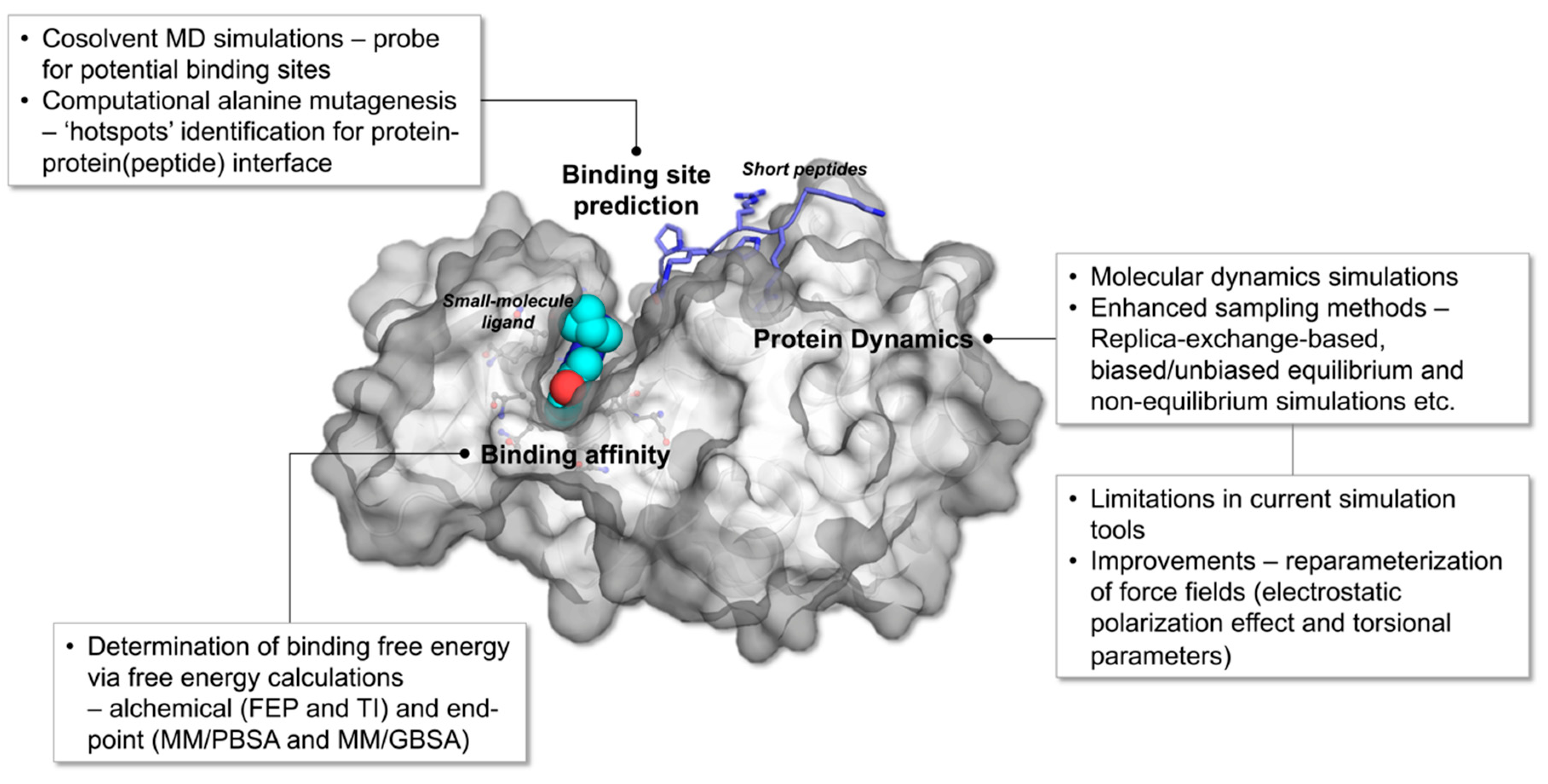
Ijms Free Full Text Advances In Molecular Dynamics Simulations And Enhanced Sampling Methods For The Study Of Protein Systems Html

Protein Corona Composition Of Poly Ethylene Glycol And Poly Phosphoester Coated Nanoparticles Correlates Strongly With The Amino Acid Composition O Nanoscale Rsc Publishing Doi 10 1039 C6nr07022a

0 comments:
Post a Comment